The following posts contain open source code and various other software developed in the Murphy Lab. If any of the software is used, please cite the relevant paper or author(s); where applicable, citations in AMA style have been provided on the page for your convenience.
Software and websites used in the lab:
- Blender, open source 3D animation
- Inkscape, open source vector graphics software for figure making
- Fiji, a distribution of ImageJ that includes plugins for scientific image analysis
- Codeacademy, programming tutorials
Terms
You may use software of this site that is linked or described on this page for any personal, commercial or educational purpose, including installing it on as many different computers as you wish. You may also copy, distribute, modify, and/or sell it under the terms of the GNU General Public License (GPL) as published by the Free Software Foundation – either version 2 of the License, or (at your option) any later version. In granting you this right, the GPL requires that the source code you distribute is itself available under the GPL. If you have questions, please contact us.
Synthetic Animated Mouse (SAM)
“We develop a 3D synthetic animated mouse based on CT scans that is actuated using animation and semi-random, joint-constrained movements to generate synthetic behavioural data with ground-truth label locations. Image-domain translation produced realistic synthetic videos used to train 2D and 3D pose estimation models with accuracy similar to typical manual training datasets. The outputs from the 3D model-based pose estimation yielded better definition of behavioral clusters than 2D videos and may facilitate automated ethological classification.” (Bolanos et al., 2021).
Links:OSF page – Includes datasets, Github link, and other resources |
 |
Software and hardware for automated mouse tapered beam test
This device can also be used for any application where the touch of a paw or tongue needs to be tracked.
Links:Ardesch DJ, Balbi M, Murphy TH. Automated touch sensing in the mouse tapered beam test using Raspberry Pi. J Neurosci Methods. 2017;291:221-226. doi:10.1016/j.jneumeth.2017.08.030 |
 |
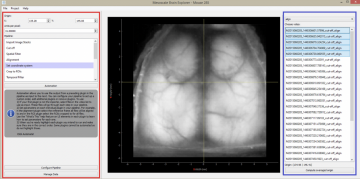
Software developed for mesoscale brain mapping, open source Python with GUI
Links:GitHub repository (see README) Haupt D, Vanni MP, Bolanos F, Mitelut C, LeDue JM, Murphy TH. Mesoscale brain explorer, a flexible python-based image analysis and visualization tool. Neurophotonics. 2017;4(3):031210. doi:10.1117/1.nph.4.3.031210 |
 |
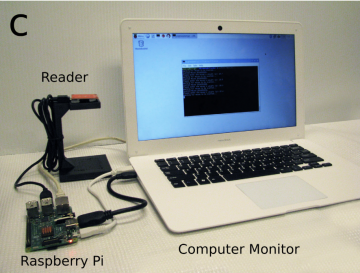
Software developed RFID tagging of mice Python
Bolaños F, LeDue JM, Murphy TH. Cost effective raspberry pi-based radio frequency identification tagging of mice suitable for automated in vivo imaging. J Neurosci Methods. 2017;276:79-83. doi:10.1016/J.JNEUMETH.2016.11.011
Links: |
 |
Neurophotonics tutorial on making connectivity diagrams from Channelrhodopsin-2 stimulated data
This package of sample data and MATLAB codes will process, filter, and average raw voltage sensitive dye images, and generate a comprehensive connectivity matrix and a network diagram, as seen in the papers below.
Links:Requirements:MATLAB (with Bioinformatics Toolbox and Brain Connectivity Toolbox) Inputs:sample data; MATLAB functions; Mask.tif (as included in .zip file) Outputs:dF/F0(%) VSD connectivity matrix; Figure of network properties as a function of threshold levels; Network diagram Lim DH, Mohajerani MH, LeDue J, Boyd J, Chen S, Murphy TH. In vivo large-scale cortical mapping using channelrhodopsin-2 stimulation in transgenic mice reveals asymmetric and reciprocal relationships between cortical areas. Front Neural Circuits. 2012;6:11. doi:10.3389/fncir.2012.00011 Lim DH, LeDue JM, Murphy TH. Network analysis of mesoscale optical recordings to assess regional, functional connectivity. Neurophotonics. 2015;2(4):041405. doi:10.1117/1.nph.2.4.041405 |
 |
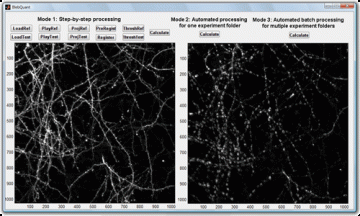
BlebQuant: Automated and quantitative analysis of blebbed dendrites
| BlebQuant is developed for automated detection and quantification of blebbing. The program is a MATLAB-based graphical user interface.
Written by Shangbin Chen Links: |
 |
Interpolate NaN
Small plugin for ImageJ to interpolate pixels with value “NaN” and “infinity” in images or image stacks. This tool is supposedly useful to apply before Gaussian or mean filtering data because it avoids the subsequent formation of filtering artifacts. Options of this plugin may be set to match the parameters of the filter applied thereafter. Written by Albrecht Sigler.
Links:Interpolate_NaN.class (compiled plugin) Interpolate_NaN.java (source code)
Seen in:
Chen S, Tran S, Sigler A, Murphy TH. Automated and quantitative image analysis of ischemic dendritic blebbing using in vivo 2-photon microscopy data. J Neurosci Methods. 2011;195(2):222-231. doi:10.1016/j.jneumeth.2010.12.018 |
 |
Dispose All Windows: ImageJ plugin
Links:Dispose_All_Windows.jar (compiled plugin) Dispose_All_Windows.java (source code) This ImageJ plugin closes all image windows and non-image windows as selected by the user. The code is based on the Dispose_All_Windows plugin by Tony Collins and J. Anthony Parker. The main plugin features should work with all common versions of ImageJ. The optional functionality of closing non-image windows requires ImageJ version 1.40 or newer. Fixed and updated Jan 5, 2013 by Albrecht Sigler. **not all information was moved over for this post |
No Image |
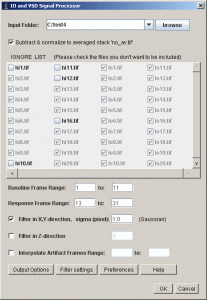
IO and VSD Processor
| Plugin for ImageJ for analysis of Intrinsic Optical Signal (IOS) or Voltage Sensitive Dye (VSD) fluorescence signal in imaging data.
Written by Albrecht Sigler LinksTo install, copy the file into the plugin folder and restart ImageJ. Detailed information about handling plugins can be found at the Plugins website of ImageJ. Requires ImageJ version 1.31v or later, whereas updating to newer ImageJ versions is recommended for performance. Open Source; feel free to modify and to distribute according to the rules of the GNU General Public License. PDF file with detailed program description and user guide.
Seen in:Harrison TC, Sigler A, Murphy TH. Simple and cost-effective hardware and software for functional brain mapping using Intrinsic Optical Signal imaging. J Neurosci Methods. 2009;182(2):211-218. doi:10.1016/j.jneumeth.2009.06.021 |
 |